Preprocess and Pipelines
Preprocess and Pipelines
scikit-learn requirements
- Numeric data
- No missing values
With real-world data:
- This is rarely the case
- We will often need to preprocess our data first
Deal with categorical features
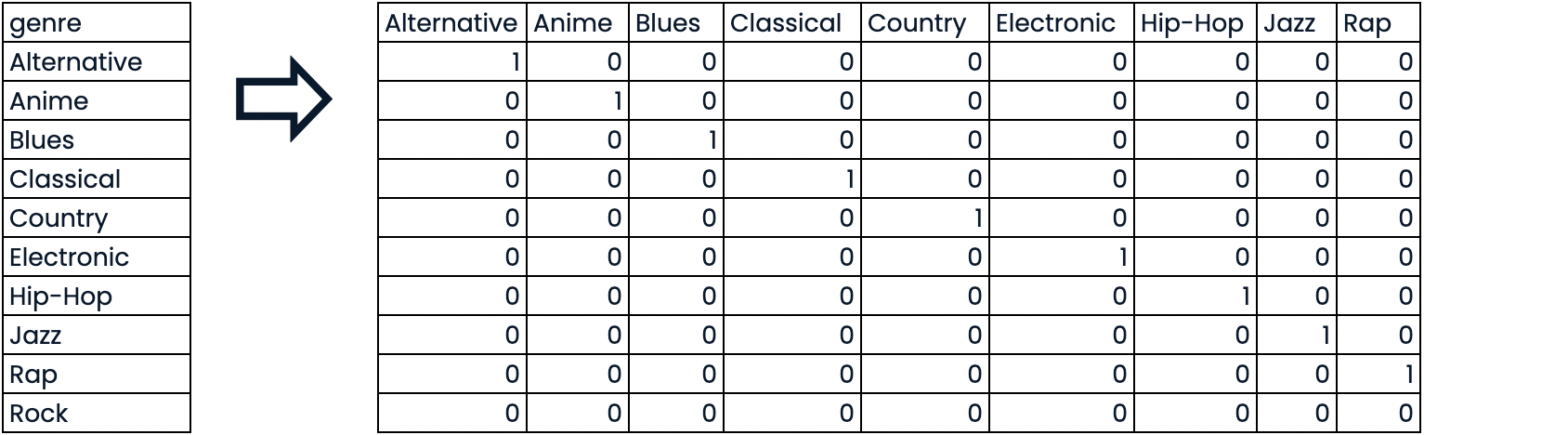
假设我们有一个包含分类特征的数据集,比如说颜色。由于这些不是数字,scikit-learn不会接受它们,我们需要将它们转换成数字特征。我们通过将该特征分割成多个二进制特征,称为虚拟变量,每个类别一个,来实现这一目标。0 意味着观察结果不是那个类别,而 1 意味着它是。
提示
适用于 X 中包含分类特征,y 是数值。
- scikit-learn will not accept categorical features by default
- Need to convert categorical features into numeric values
- Convert to binary features called dummy variables
- 0: Observation was NOT that category
- 1: Observation was that category
提示
删除 "Rock "一栏。如果我们不这样做,我们就会重复信息,这对某些模型来说可能是个问题。
为了创建虚拟变量,我们可以使用
- scikit-learn: OneHotEncoder()
- pandas: get_dummies()
Example: Music dataset
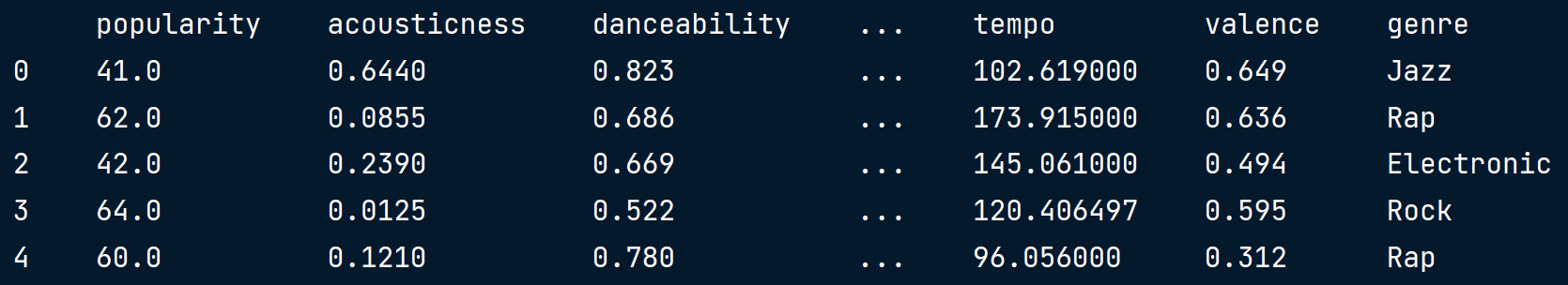
EDA w/ categorical feature
- popularity : Target variable
- genre : Categorical feature
print(music.info())
Encoding dummy variables
import pandas as pd
music_df = pd.read_csv('music.csv')
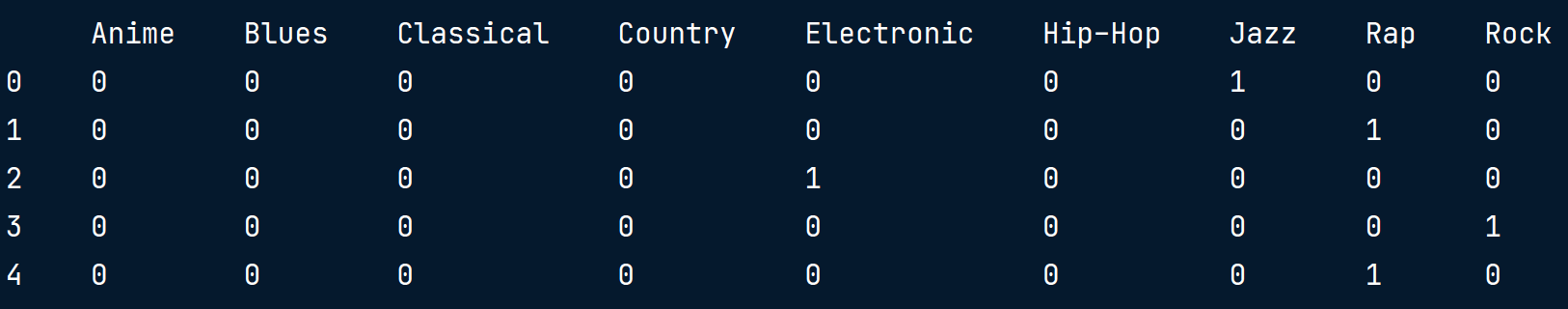
# 由于我们只需要保留10个二进制特征中的9个,我们可以将drop_first参数设置为True。
music_dummies = pd.get_dummies(music_df["genre"], drop_first=True)
print(music_dummies.head())
music_dummies = pd.concat([music_df, music_dummies], axis=1)
music_dummies = music_dummies.drop("genre", axis=1)
如果DataFrame只有一个分类特征,我们可以传递整个DataFrame,从而跳过组合变量的步骤。如果我们不指定列,新的DataFrame的二进制列将有原始特征名称的前缀,所以它们将以 genre_ 开始。注意原来的流派列会被自动删除。一旦我们有了虚拟变量,我们就可以像以前一样拟合模型。
music_dummies = pd.get_dummies(music_df, drop_first=True)
print(music_dummies.columns)

Linear regression
from sklearn.model_selection import cross_val_score, KFold
from sklearn.linear_model import LinearRegression
X = music_dummies.drop("popularity", axis=1).values
y = music_dummies["popularity"].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
kf = KFold(n_splits=5, shuffle=True, random_state=42)
linreg = LinearRegression()
linreg_cv = cross_val_score(linreg, X_train, y_train, cv=kf, scoring="neg_mean_squared_error")
print(np.sqrt(-linreg_cv))
[8.15792932, 8.63117538, 7.52275279, 8.6205778, 7.91329988]
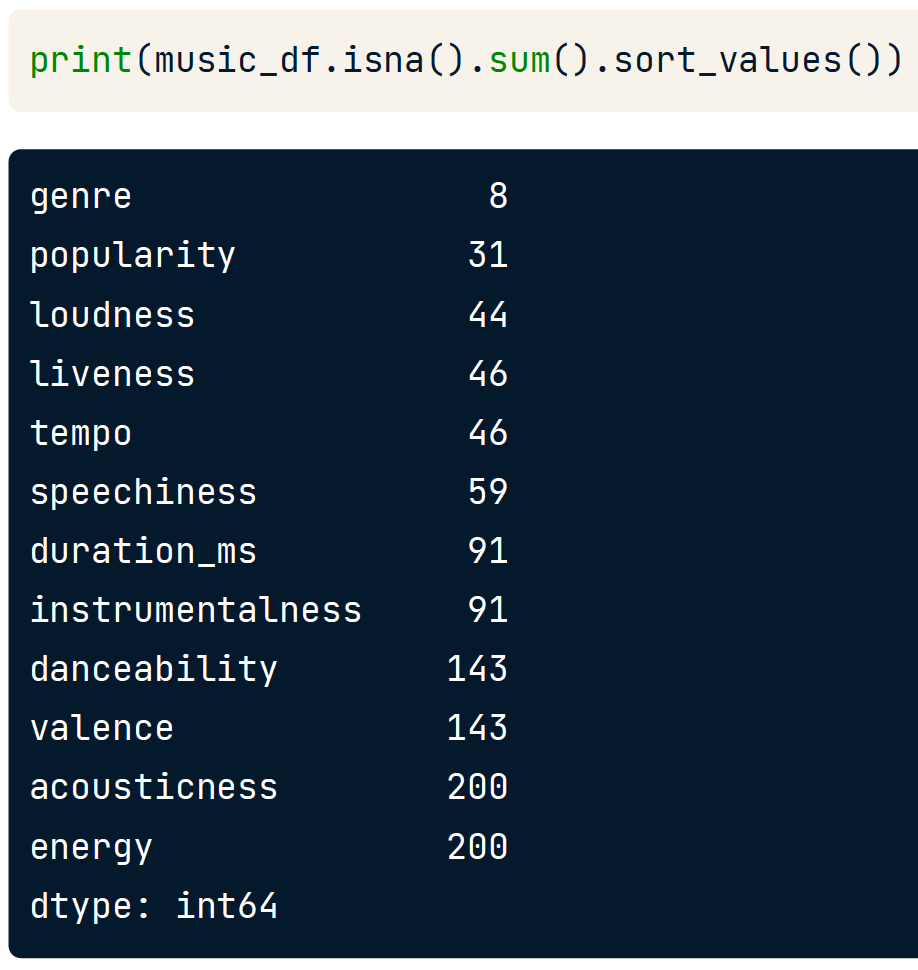
Handling missing data
No value for a feature in a particular row, This can occur because:
- There may have been no observation
- The data might be corrupt
We need to deal with missing data
Dropping missing data
一个常见的方法是移除占所有数据5%以下的缺失观测值。为了做到这一点,我们使用pandas的dot-dropna方法,将一个缺失值小于5%的列的列表传递给subset参数。如果在我们的子集列中存在缺失值,那么整行都会被删除。重新检查DataFrame,我们看到更少的缺失值。
Imputing values
代入法「Imputing」 —— 利用受试者的专业知识,用有根据的猜测取代缺失的数据
- Common to use the mean, Can also use the median, or another value
- For categorical values, we typically use the most frequent value - the mode
- 首先必须拆分我们的数据,以避免数据泄漏
# Imputation with scikit-learn
from sklearn.impute import SimpleImputer
X_cat = music_df["genre"].values.reshape(-1, 1)
X_num = music_df.drop(["genre", "popularity"], axis=1).values
y = music_df["popularity"].values
X_train_cat, X_test_cat, y_train, y_test = train_test_split(X_cat, y, test_size=0.2, random_state=12)
X_train_num, X_test_num, y_train, y_test = train_test_split(X_num, y, test_size=0.2, random_state=12)
imp_cat = SimpleImputer(strategy="most_frequent")
X_train_cat = imp_cat.fit_transform(X_train_cat)
X_test_cat = imp_cat.transform(X_test_cat)
imp_num = SimpleImputer()
X_train_num = imp_num.fit_transform(X_train_num)
X_test_num = imp_num.transform(X_test_num)
X_train = np.append(X_train_num, X_train_cat, axis=1)
X_test = np.append(X_test_num, X_test_cat, axis=1)
Imputers are known as transformers
Imputing within a pipeline
from sklearn.pipeline import Pipeline
music_df = music_df.dropna(subset=["genre", "popularity", "loudness", "liveness", "tempo"])
music_df["genre"] = np.where(music_df["genre"] == "Rock", 1, 0)
X = music_df.drop("genre", axis=1).values
y = music_df["genre"].values
steps = [("imputation", SimpleImputer()), ("logistic_regression", LogisticRegression())]
pipeline = Pipeline(steps)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
pipeline.fit(X_train, y_train)
pipeline.score(X_test, y_test)
0.7593582887700535
注意
请注意,在一个管道中,除了最后一步外,每一步都必须是一个转化器。
Centering and scaling
许多机器学习模型使用某种形式的距离来告知它们,所以如果我们有远大于尺度的特征,它们会不成比例地影响我们的模型。例如,KNN在进行预测时明确地使用距离。出于这个原因,我们实际上希望特征在一个类似的尺度上。为了实现这一点,我们可以将我们的数据规范化或标准化,通常被称为缩放和居中「Normalizing or standardizing (scaling and centering)」。
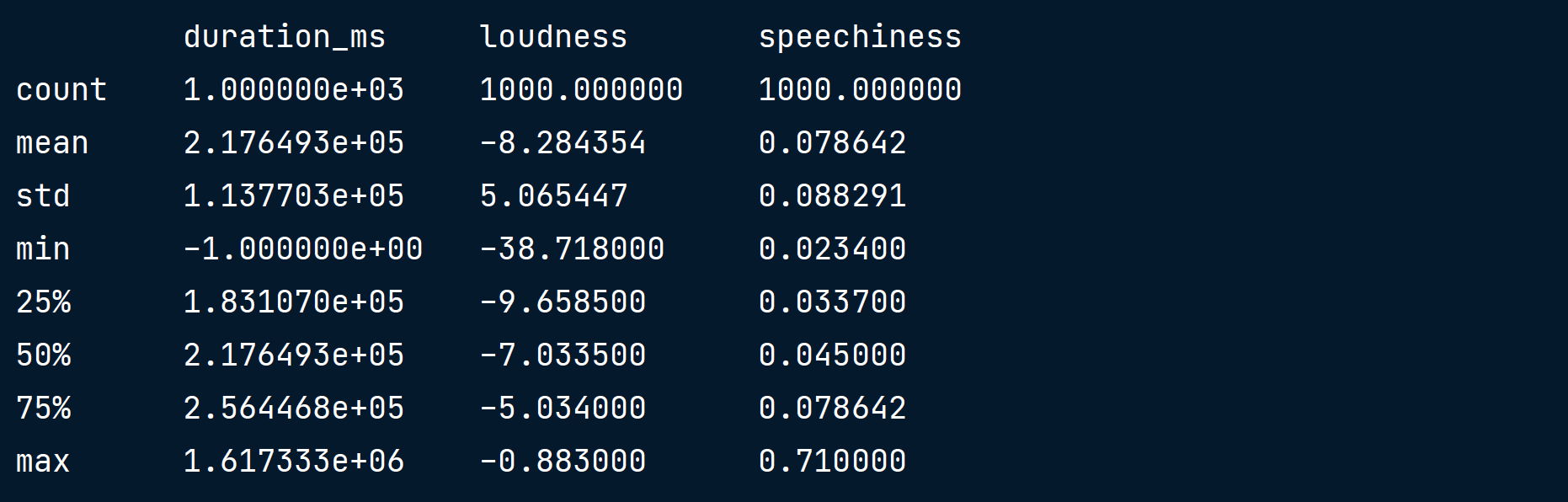
print(music_df[["duration_ms", "loudness", "speechiness"]].describe())
How to scale our data
- 减去平均值并除以方差
- 所有 features 都以 0 为中心,方差为 1
- This is called standardization
- 也可以减去最小值并除以 range
- 最小为0,最大为1
- 也可以归一化,使数据范围从-1到+1。
请参阅 scikit - 学习文档以获取更多详细信息
Scaling in scikit-learn
from sklearn.preprocessing import StandardScaler
X = music_df.drop("genre", axis=1).values
y = music_df["genre"].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
# 对比缩放前后
print(np.mean(X), np.std(X))
print(np.mean(X_train_scaled), np.std(X_train_scaled))
19801.42536120538, 71343.52910125865
2.260817795600319e-17, 1.0
Scaling in a pipeline
steps = [('scaler', StandardScaler()), ('knn', KNeighborsClassifier(n_neighbors=6))]
pipeline = Pipeline(steps)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=21)
knn_scaled = pipeline.fit(X_train, y_train)
y_pred = knn_scaled.predict(X_test)
print(knn_scaled.score(X_test, y_test))
0.81
CV and scaling in a pipeline
from sklearn.model_selection import GridSearchCV
steps = [('scaler', StandardScaler()), ('knn', KNeighborsClassifier())]
pipeline = Pipeline(steps)
parameters = {"knn__n_neighbors": np.arange(1, 50)}
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=21)
cv = GridSearchCV(pipeline, param_grid=parameters)
cv.fit(X_train, y_train)
y_pred = cv.predict(X_test)
print(cv.best_score_)
print(cv.best_params_)
0.8199999999999999
{'knn__n_neighbors': 12}